 News
News
With the exponential growth of digital data production, conventional storage technologies face increasing challenges related to scalability, durability, and environmental impact. DNA has emerged as a promising alternative storage medium due to its exceptional density, longevity, and sustainability. In this context, the JPEG Committee (ISO/IEC JTC 1/SC 29/WG 1) has developed the JPEG DNA standard (ISO/IEC 25508), the first specification for image coding on synthetic DNA, which is currently in its final stages of standardization. The goal of the standard is to enable the efficient and robust representation of digital images using DNA as a storage medium, while addressing challenges such as biochemical constraints and high error rates. This paper provides an overview of the JPEG DNA standard, describing in detail its codec-agnostic core coding system able to encode not only images but also arbitrary binary data, and presenting a performance comparison with state-of-the-art alternatives. In particular, the codec is observed to be able to encode data at 1.83 bits/nt without producing homopolymers or repeated patterns and keeping a balanced content of C and G nucleotides. In addition, a discussion is provided on ongoing developments and remaining challenges in the standardization process, including a wide discussion of the performance of the JPEG DNA standard in the presence of noise.
The exponential increase in storage demand and low lifespan of data storage devices has resulted in long-term archival and preservation emerging as critical bottlenecks in data storage. In order to meet this demand, researchers are now investigat- ing novel forms of data storage media. The high density, long lifespan and low energy needs of synthetic DNA make it a promising candidate for long-term data archival. However, current DNA data storage technologies are facing challenges with respect to cost (writing data to DNA is expensive) and reliability (reading and writing data is error prone). Thus, data compression and error correction are crucial to scale DNA storage. Additionally, the DNA molecules encoding several files are very often stored in the same place, called an oligo pool. For this reason, without random access solutions, it is relatively impractical to decode a specific file from the pool, because all the oligos from all the files need to first be sequenced, which greatly deteriorates the read cost. This paper introduces PIC-DNA–a novel JPEG2000-based progressive image coder adapted to DNA data storage. This coder directly includes a random access process in its coding system, allowing for the retrieval of a specific image from a pool of oligos encoding several images. The progressive decoder can dynamically adapt the read cost according to the user’s cost and quality constraints at decoding time. Both the random access and progressive decoding greatly improve on the read-cost of image coders adapted to DNA.
The increase in storage demand and low lifespan of conventional data storage media has transformed long-term archival and preservation into key bottlenecks for the data storage industry. Thus, researchers are now investigating innovative data storage media techniques. DNA molecules, with their high density, long lifespan and low energy needs, are promising candidates for alternative long- term data archival systems.
However, current DNA data storage technologies are facing challenges with respect to cost (reading and writing on DNA is expensive) and reliability (reading and writing data is error prone). Thus, data compression and error correction are crucial to scale DNA storage and make it technologically and economically viable. Additionally, the DNA molecules encoding different files are very often stored in the same place, called an oligo pool. For this reason, without random access solutions, it is relatively impractical to decode a specific file from the pool, because all the oligos from all the files need to first be sequenced, which greatly deteriorates the read cost.
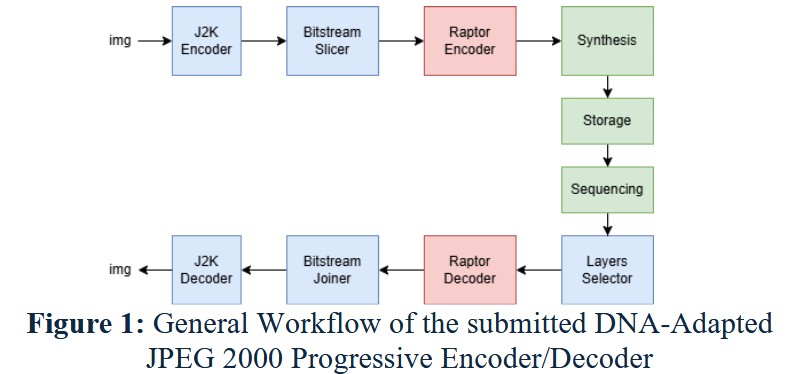
This paper introduces a solution (Fig. 1) to efficiently encode and store images into DNA molecules, that aims at reducing the read cost necessary to retrieve a resolution-reduced version of an image. This image storage system is based on the Progressive Decoding Functionality of the JPEG2000 codec and can be adapted for any other codec that enables a progressive decoding function. Each resolution layer is encoded into a set of oligos using the Raptor code [1] provided in the JPEG DNA VM software, with primers specific to the resolution layer attached to them. Depending on the desired resolution to be read, the set of oligos to be sequenced and decoded is adjusted accordingly. These oligos will be selected, augmented and sequenced through a PCR process run with the layer specific primers. The ReadUntil functionality of the Nanopore Sequencer can replace the PCR runs. It provides a system to reject at sequencing time the oligos that do not match a given template. This template can be dynamically modified during sequencing, allowing for better automation of the whole layer access process.
References:
[1] Lazzarotto, D. et al., "Technical description of the EPFL submission to the JPEG DNA CfP",
jpeg.org, 2023
I am very happy to join the team of Pr. Raja Appuswamy, in the Data Science department, to work on image compression based on AI models. An application to DNA storage is also considered. This postdoc is funded by the French Government, under the PEPR Molexcularxiv Project fund.
Coding algorithms for long-term storage of digital images on synthetic DNA molecules
Abtract
The current digital world is facing a number of issues, some of them linked to the amount of data that is being stored. The current technologies available as an offer to store data are not enough to store the totality of the storage demand. For this reason, new data storage technologies have to be developed. DNA molecules are one of the candidates available for novel data storage methods. The long lifespan of these molecules make it a good fit for the archival of data that is rarely accessed but needs to be stored for long periods of time. This data, often called “cold”, represents approximately 80% of the data in our digital universe. But DNA uses 4 symbols (A,C,G and T) to encode data against the usual binary code (0,1). For this reason, storing data into DNA requires a specific encoding system capable of translating a binary data stream into a quaternary data stream. In this thesis we will focus on new encoding methods from the Deep Learning state of the art, and we will adapt those methods for the encoding, decoding, compression and decompression of images on synthetic DNA.
Jury
- Aline Roumy, Research Director, INRIA, Rennes
- Eitan Yaakobi, Research Director, Technion, Haifa
- Thomas Heinis, Associate Professor, Imperial College London
- Athanassios Skodras, Professor, University of Patras
- Raja Appuswamy, Assistant Professor, EURECOM, Sophia Antipolis
- Dominique Lavenier, Research Director, CNRS, IRISA, Rennes
Over the past years, the ever-growing trend on data storage demand, more specifically for "cold" data (i.e. rarely accessed), has motivated research for alternative systems of data storage. Because of its biochemical characteristics, synthetic DNA molecules are now considered as serious candidates for this new kind of storage. This paper introduces a novel arithmetic coder for DNA data storage, and presents some results on a lossy JPEG 2000 based image compression method adapted for DNA data storage that uses this novel coder. The DNA coding algorithms presented here have been designed to efficiently compress images, encode them into a quaternary code, and finally store them into synthetic DNA molecules. This work also aims at making the compression models better fit the problematic that we encounter when storing data into DNA, namely the fact that the DNA writing, storing and reading methods are error prone processes. The main take away of this work is our arithmetic coder and it's integration into a performant image codec.

Multiple Description Coding (MDC) is an error-resilient source coding method designed for transmission over noisy channels. We present a novel MDC scheme employing a neural network based on implicit neural representation. This involves overfitting the neural representation for images. Each description is transmitted along with model parameters and its respective latent spaces. Our method has advantages over traditional MDC that utilizes auto-encoders, such as eliminating the need for model training and offering high flexibility in redundancy adjustment. Experiments demonstrate that our solution is competitive with autoencoder-based MDC and classic MDC based on HEVC, delivering superior visual quality.
Over the past years, the ever-growing trend on data storage demand, more specifically for "cold" data (rarely accessed data), has motivated research for alternative systems of data storage. Because of its biochemical characteristics, synthetic DNA molecules are now considered as serious candidates for this new kind of storage. This paper presents some results on lossy image compression methods based on convolutional autoencoders adapted to DNA data storage, with synthetic DNA-adapted entropic and fixed-length codes. The model architectures presented here have been designed to efficiently compress images, encode them into a quaternary code, and finally store them into synthetic DNA molecules. This work also aims at making the compression models better fit the problematics that we encounter when storing data into DNA, namely the fact that the DNA writing, storing and reading methods are error prone processes. The main take aways of this kind of compressive autoencoder are our latent space quantization and the different DNA adapted entropy coders used to encode the quantized latent space, which are an improvement over the fixed length DNA adapted coders that were previously used.
The JPEG Committee has been exploring coding of images in quaternary representations particularly suitable for image archival on DNA storage. The scope of JPEG DNA is to create a standard for efficient coding of images that considers biochemical constraints and offers robustness to noise introduced by the different stages of the storage process that is based on DNA synthetic polymers.
At the 100th JPEG meeting, “Additions to the JPEG DNA Common Test Conditions version 2.0”, was produced which supplements the “JPEG DNA Common Test Conditions” by specifying a new constraint to be taken into account when coding images in quaternary representation. In addition, the detailed procedures for evaluation of the pre-registered responses to the JPEG DNA Call for Proposals were defined.
Furthermore, the next steps towards a deployed high-performance standard were discussed and defined. In particular, it was decided to request for the new work item approval once a Committee Draft stage has been reached.
The JPEG-DNA AHG has been re-established to work on the preparation of assessment and crosschecking of responses to the JPEG DNA Call for Proposals until the 101st JPEG meeting in October 2023.
Réunion: Stockage de données numériques dans de l'ADN synthétique - Grandes avancées et défis à relever
Org: Marc Antonini (I3S CNRS UCA), Elsa Dupraz (IMT-Atlantique), Dominique Lavenier (IRISA)
Présentation: Constrained and robust image coding for molecular storage
L'explosion des données est l'un des plus grands défis de l'évolution numérique. La demande de stockage augmente à un rythme tel qu'elle ne peut rivaliser avec les capacités réelles des appareils. Selon les prévisions, l'univers numérique devrait atteindre plus de 180 zettaoctets d'ici 2025, tandis que 80 % des données sont rarement consultées (données "froides"), mais méritent d'être archivées à long terme pour mémoire de l'humanité (photographies, films, code informatique, connaissances scientifiques, etc.). Dans le même temps, les dispositifs de stockage classiques ont une durée de vie limitée à 10 ou 20 ans et doivent être fréquemment remplacés pour garantir la fiabilité des données, un processus coûteux en termes d'argent et d'énergie. Des études récentes ont montré qu'en raison de ses propriétés biologiques, l'ADN est un candidat très prometteur pour l'archivage à long terme de données numériques "froides" pendant des siècles. Le stockage de données sous la forme de molécules d'ADN nécessite de coder les informations dans un flux quaternaire composé des symboles A, C, T et G (les fameux nucléotides), tout en respectant des contraintes strictes liées aux processus biochimiques associés. De plus, ce support de stockage introduit des erreurs non conventionnelles de types insertions et deletions que les méthodes classiques de correction d'erreurs ne savent pas traiter. Des travaux pionniers ont d'ores et déjà proposé différents algorithmes pour le codage et la protection des données stockées dans de l'ADN, laissant cependant encore la place à de nombreux défis à relever.
L'objectif de cette journée est de faire le point sur les avancées technologiques et les grands défis à relever dans ce domaine du stockage moléculaire, en mettant en avant les problématiques liées au traitement du signal et des images ainsi que de la théorie des codes correcteurs et du codage source/canal conjoint. La journée débutera par deux tutoriels d'introduction au sujet, avant de se poursuivre par des exposés plus techniques sur les sujets précédents.
Over the past years, the ever-growing trend on data storage demand, more specifically for "cold" data (i.e. rarely accessed), has motivated research for alternative systems of data storage. Because of its biochemical characteristics, synthetic DNA molecules are considered as potential candidates for a new storage paradigm. Because of this trend, several coding solutions have been proposed over the past years for the storage of digital information into DNA. Despite being a promising solution, DNA storage faces two major obstacles: the large cost of synthesis and the noise introduced during sequencing. Additionally, this noise increases when biochemically defined coding constraints are not respected: avoiding homopolymers and patterns, as well as balancing the GC content. This paper describes a novel entropy coder which can be embedded to any block-based image-coding schema and aims to robustify the decoded results. Our proposed solution introduces variability in the generated quaternary streams, reduces the amount of homopolymers and repeated patterns to reduce the probability of errors occurring. In this paper, we integrate the proposed entropy coder into four existing JPEG-inspired DNA coders. We then evaluate the quality-in terms of biochemical constraints-of the encoded data for all the different methods.
The exponentially increasing demand for data storage has been facing more and more challenges during the past years. The energy costs that it represents are also increasing, and the availability of the storage hardware is not able to follow the storage demand's trend. The short lifespan of conventional storage media-10 to 20 years-forces the duplication of the hardware and worsens the situation. The majority of this storage demand concerns "cold" data, data very rarely accessed but that has to be kept for long periods of time. The coding abilities of synthetic DNA, and its long durability (several hundred years), make it a serious candidate as an alternative storage media for "cold" data. In this paper, we propose a variable-length coding algorithm adapted to DNA data storage with improved performance. The proposed algorithm is based on a modified Shannon-Fano code that respects some biochemichal constraints imposed by the synthesis chemistry. We have inserted this code in a JPEG compression algorithm adapted to DNA image storage and we highlighted an improvement of the compression ratio ranging from 0.5 up to 2 bits per nucleotide compared to the state-of-the-art solution, without affecting the reconstruction quality.








